AI will map proteins and increase understanding of how cells work
Artificial intelligence (AI) and deep learning are becoming increasingly important to map the proteins in our cells and how they interact with each other. In a new project, the researchers will develop tools to increase understanding of how cells work.

Within the life sciences, a major transformation has taken place in recent decades. From having been "data poor" there is now great access to research data. In recent years, there has also been a revolution in the development of deep learning methods that make it possible to use large amounts of data.
By combining the latest advances in these two fields, opportunities are created to revolutionise medical treatments. There are already examples of such progress, mainly in research on predicting protein structures. The deep learning tool AlphaFold from DeepMind is perhaps the clearest example. In the summer of 2021, two articles were published about AlphaFold and DeepMind released the code freely to all researchers. This started a process where hundreds of researchers have now used the code in different ways and resulted in many scientific publications.
30 million for research about cells
A research group led by Arne Elofsson, professor of bioinformatics at Stockholm University and active at the Department of Biochemistry and Biophysics and SciLifeLab, has now been awarded a research grant of 30 million by the Knut and Alice Wallenberg Foundation for research into the molecular components of the cell. The group also includes researchers at Karolinska Institutet and KTH. The researchers will use deep learning methods to identify all protein forms present in a cell, as well as their interactions with each other. The 20,000 genes found in humans give rise to many more protein forms through splicing and posttranslational modifications. Exactly how many protein forms exist and which are important is still largely unknown.
“Within the project, we hope to be able to map most of the protein interactions found in a human cell. To achieve this, we will need to develop new methods based on artificial intelligence and exploit new information from large-scale experiments. We hope that the project will lead to the development of new tools that can be used by everyone, as well as to an increased understanding of how a cell works,” says Arne Elofsson.
Even more accurate models of protein complexes
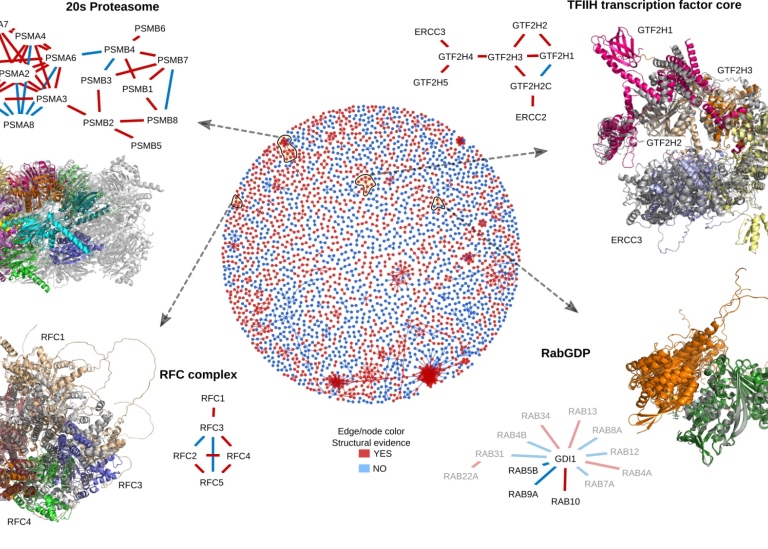
He and his colleagues have already used AlphaFold and developed new methods for studying interactions between proteins and improved methods for predicting the structure of large protein complexes. The protein forms they hope to identify will now be used to build even more accurate models of protein complexes. To test predictions and create more experimental data, they will also use mass spectrometry. It is a technique that can quickly provide information on how proteins fold and interact.
To be able to model all the proteins and their interactions in a human cell, however, even better deep learning methods, more experimental data and access to a lot of data capacity are needed. Here, the researchers will make use of the rapid development that has taken place in open software. The research group has already developed its own open software and will continue to do so.
Read article: Four large Wallenberg grants to Stockholm University
Last updated: October 18, 2022
Source: Communications Office

