New Genomics Tool enables increased understanding of the liver
Researchers at the Department of Molecular Biosciences, the Wenner-Gren Institute, Stockholm University, have applied Spatial Transcriptomics, ST to study the spatial organization of the liver.

“In our study, we applied this method to study total gene expression of small numbers of cells across tissue space in liver tissue sections,” says Dr Johan Ankarklev, Principal investigator at MBW, Department of Molecular Biosciences, the Wenner-Gren Institute, Stockholm University, who has led the study in a collaboration with Prof Joakim Lundeberg at the Royal Institute of Technology, and several other research groups at Stockholm University, the Royal Institute of Technology, the Karolinska Institute and SciLifeLab.
The liver is a highly spatially heterogenous tissue, which fulfills pivotal functions such as metabolic regulation, including glucose homeostasis and cholesterol or protein synthesis as well as detoxification. While single-cell transcriptional profiling has greatly advanced our understanding of the spatial liver transcriptome in recent years, a comprehensive description of spatially defined global gene expression across whole tissue sections has been lacking.
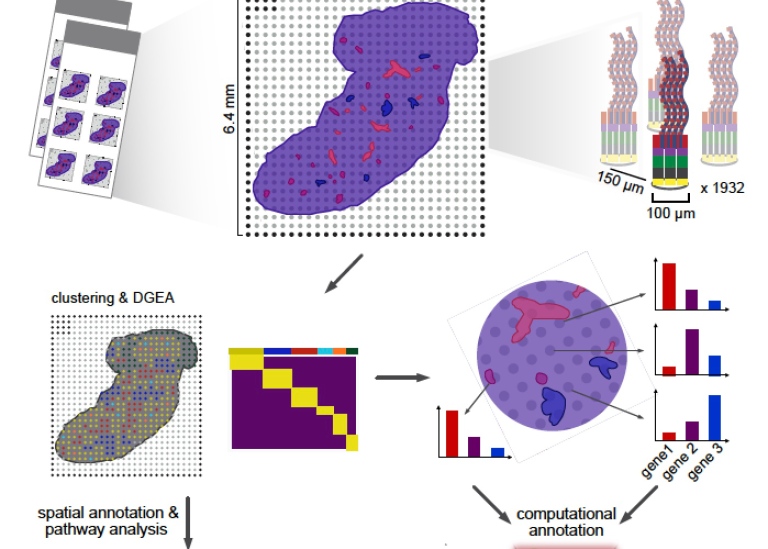
The basis of the technology Spatial Transcriptomics, ST, is a glass-slide array with barcoded capture regions (spots) where each spot has the ability of capturing mRNA transcripts from the tissue that is placed directly on top of it. The method then combines imaging data with transcriptome data from the same tissue, which opens up for a new understanding of the spatial organization of the liver.
“We are excited to show in our data how spatial transcriptomic technologies can be used to describe spatial gene expression patterns in the liver, which indicates its future impact for studies of liver function, development, and regeneration as well as its potential in pre-clinical and clinical pathology, and of course infection biology,” says Dr Ankarklev.
The researchers have also generated novel computational approaches that enable transcriptional gradient measurements between tissue structures across the entire tissue.
For instance, they have been able to measure division of metabolic activity (zonation) across the liver, at high resolution and in its true tissue context. The study was performed on tissue sections from mouse livers but the method is applicable for use on human liver tissue as well.
“Finally, I want to highlight the great work by Franziska Hildebrandt, a PhD student in my lab and the fantastic team effort by our collaborators, whose diverse expertise made this study possible”, says Johan Ankarklev.
Last updated: December 2, 2021
Source: Communication Office

