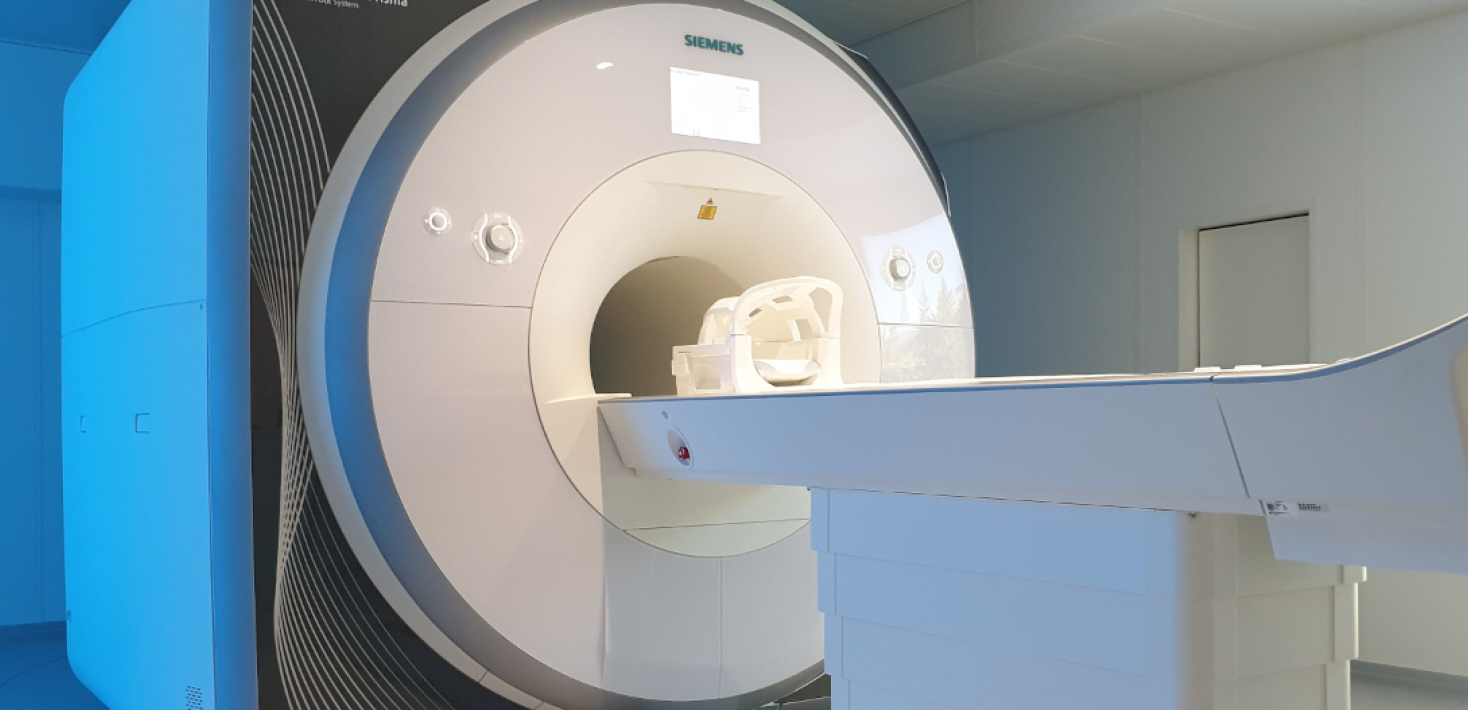
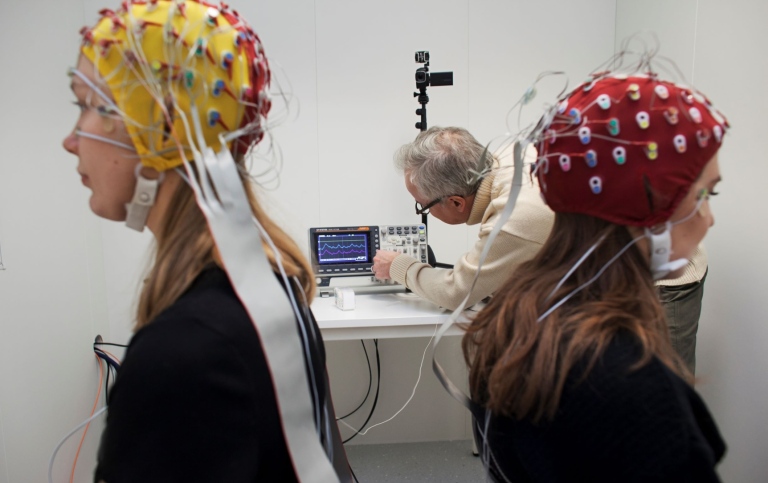
SUBIC – Stockholm University Brain Imaging Centre
SUBIC is a multidisciplinary infrastructure dedicated to research on brain structure and function, as well as other fields benefiting from imaging micro structures
SUBIC’s infrastructure supports Swedish research with state of the art imaging technologies, including MRI, EEG, TMS, X-ray microscopy. SUBIC's equipment is valuable for fundamental and applied academic research within the humanities, social sciences, law and natural sciences.
To receive information from SUBIC please register below: