Mating-type switching has evolved on at least three independent occasions in yeasts. S. cerevisiae, K. lactis and S. pombe all have distinct mechanisms for switching. In common for all three species is that cryptic mating type loci act as donors of mating type information, which is copied from the cryptic (non-expressed) loci into the expressed Mating type (MAT) locus. In K. lactis, the mechanism and regulation of switching is only partly understood, motivating further studies of this organism.
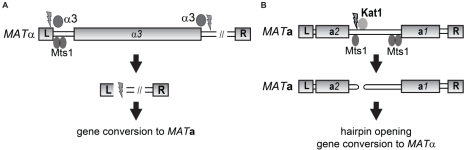
We have found that two different proteins mediate switching from MATa to MATα (Kat1) and MATα to MATa (α3) respectively (See figure). Remarkably, both of these proteins are domesticated transposases, linking selfish DNA with mating-type switching. The translation of one of the transposase genes requires a programmed -1 frameshift. Moreover, we have evidence for that this frameshift is important to prevent excessive activation of the transposase and hence that the cell population switches completely to the MATa genotype. The Kat1 transposase generates hairpin-capped breaks in DNA, and we are studying how the cells open and repair this hairpin-capped DSB. We have also found that mating pheromones inhibit switching and we suggest that this is a way for K. lactis to promote outcrossing at the expense of inbreeding.
Through database mining and large scale sequencing efforts we have obtained the genome sequence of four yeast species (K. aestuarii, K.dobzhanskii, K. marxianus and K. wickerhamii) that are the closest known relatives of K. lactis. This enables us to use comparative genomics to learn more about the mechanism and regulation of mating-type switching.
Links
Saccharomyces genome database – a comprehensive knowledge base of Baker’s yeast http://www.yeastgenome.org/
Genolevures a site for genomic exploration of other ascomycetes http://www.genolevures.org/
An excellent lecture on DNA repair/recombination
http://www.youtube.com/watch?v=UzXcq2h0VCk




