1. Gene network controlled by Notch signaling and cell-type specific transcription factors (TFs) in Drosophila periphery nervous system (PNS).
The Drosophila adult peripheral nervous system (PNS) comprises mechano-sensory organs located on the body surface of adult flies. Each organ contains 4-5 terminal cells that arise from iterative asymmetric divisions of a sensory organ precursor (SOP). This system has been extremely fruitful in identifying novel regulators and mechanisms in cell fate specification. For example, decades of genetic studies revealed that Notch signaling and cell-type specific TFs collaborate to specify PNS fates. However, much remains to be understood regarding the regulation of Notch signaling, the functional interplay of Notch and cell-specific TFs, and how high-level TFs execute their function via target networks. To address these questions, our work combines sophisticated genetics possible in Drosophila, detailed molecular and biochemical studies of individual factors and loci, and high-throughput genomic approaches.

2. Gene regulation in murine neural stem cell (NSC) maintenance and differentiation.
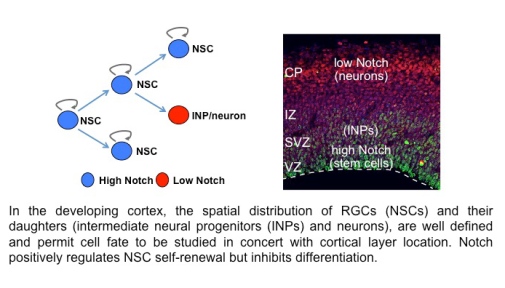
As in the fly PNS, complex lineages generate cellular diversity in the mammalian nervous system. In the developing brain and adult NSC niche, neural stem cells (NSCs) divide symmetrically to self-renew, or divide asymmetrically yielding a daughter that will eventually lead to a differentiated cell(s). Notch signaling promotes NSC self-renewal, however, very little is known about how its activity is modulated and what downstream effectors are involved. Interestingly, many homologs of Drosophila PNS TFs are also cell fate regulators in the murine nervous system. We aim to understand how the interplay between Notch signaling and conserved neural TFs control NSC self-renewal and differentiation.