Trying to understand how specific genetic variants affect whole phenotypes is challenging. We study the underlying molecular biology for phenotypes of interest, working with methylation sequencing, RNA-Seq analyses of allele specific variation, as well as CRISPR-Cas9 manipulation of candidate genes to investigate the causality of specific mutations in adaptation.
Adaptation to Environmental Stress

We live in a world where the pace of environmental change poses serious threats to biodiversity. To avoid extinction when environments deteriorate, species must evolve to adapt to the new or extreme conditions. Here, we use the microbial model system Saccharomyces yeast to understand the genomic and transcriptomic basis of adaptation, such as which and how many loci are involved in adaptation to environmental stress, and how these loci interact. Yeast genomes are easily manipulated and well suited to investigate the causality of specific mutations in adaptation.
Principal Investigator: Rike Stelkens
Contributing Researchers: Ciaran Gilchrist
Structural genomic changes and aneuploidy
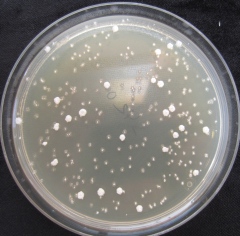
Aneuploidy is the loss or gain of extra chromosomes. Aneuploidy and other large chromosomal changes are common in crosses between divergent genomes and may provide a natural route for adaptation to changing environments. We use Saccharomyces yeast, experimental evolution, and long-read sequencing to understand the evolutionary significance of ploidy changes and chromosomal rearrangements.
Principal Investigator: Rike Stelkens
Contributing Researchers: Ciaran Gilchrist
Population genomics of butterflies
Understanding the evolutionary history of butterflies, from their origins to how populations have become adapted to their local conditions, is a central goal of our lab. We use a range of genomic based approaches, beginning with assembling high quality genomes. Onto this we then place samples of population variation from regions differening in their environmental conditions, such as from Spain and northern Sweden. Using this data we can then work to reconstruct the demographic history of populations, identify which regions may have undergone strong selective sweeps, as well as investigate specfici functional categories of genes to ask if they are showing strong signals of local adaptation compared to the rest of the genome. We also investigate the recombination dynamics within and between species, investigating how this affects the landscape of genetic variation compared to other insects.
Principal Investigators: Christopher Wheat, Karl Gotthard, Christer Wiklund, Soren Nylin
Contributing Researchers: Christen Bossu, Jason Hill, Lisa Fors, Naomi Pruisscher Keehnen, Peter Pruisscher, Ramprasad Neethiraj
Diapause induction in butterflies

Diapause is a life history strategy allowing individuals to arrest development until favourable conditions return, and it is commonly induced by shortened day-length that is latitude specific for local populations. Although understanding the evolutionary dynamics of a threshold trait like diapause induction provides insights into the adaptive process and the adaptive potential of populations, the genetic mechanism of variation in photoperiodic induction of diapause is not well understood. Our group investigates the genomic architecture of latitudinal variation in diapause induction and the selection dynamics acting upon it, in the speckled wood butterfly, as well as the green-veined white.
Principal Investigators: Christopher Wheat, Karl Gotthard, Christer Wiklund, Soren Nylin
Contributing Researchers: Peter Pruisscher, Philipp Lehmann, Olle Lindestad
Butterfly wing coloration

Butterflies being cold-blooded organisms rely on their environment to a large extent for their heat/energy requirements to perform essential life functions such as flight and oviposition. The ornamental wing colour patterns seen in butterflies has been shown to play an important role in the gain/loss of heat (i.e., thermoregulation), and thereby flight and oviposition. In our group, we are interested in understanding the evolution of wing coloration in the female butterflies of the green-veined white (Pieris napi), which exhibits geographic variation in wing melanization. Females in populations of Pieris napi napi, which occur across Europe in warm temperatures, have predominantly white upper forewings, whereas females of Pieris napi adalwinda, found in colder habitats at higher altitudes and latitudes of Switzerland and Sweden, respectively, have extremely dark forewings. Using a range of omic tools we are investigating the genetic basis of this trait.
Principal Investigators: Christopher Wheat, Soren Nylin
Contributing Researcher: Ramprasad Neethiraj
Butterfly Immunity
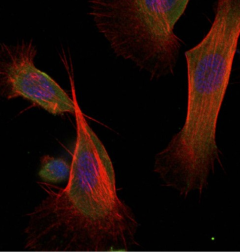
Insects rely on their innate immune system to successfully mediate complex interactions with their microbiota, as well as the microbes present in the environment. Previous work has shown that components of the canonical immune genes evolve rapidly and have evolutionary characteristics originating from interactions with fast evolving microorganisms. However, there is little understanding of how immune performance varies among populations, and we have a poor understanding of the microevolutionary dynamics acting upon these genes. In our group we both quantify the immune performance among populations of the butterfly Pieris napi. In addition, we use genomic tools to quantify microevolutionary dynamics acting on the immune system across a range of additional species.
Principal Investigators: Christopher Wheat, Soren Nylin
Contributing Researchers: Lisa Fors, Naomi Pruisscher Keehnen
Coevolutionary dynamics between butterflies and their hostplants

Coevolutionary interactions are thought to have spurred the evolution of key innovations and driven the diversification of much of life on Earth. However, the genetic and evolutionary basis of the innovations that facilitate such interactions remains poorly understood. We have previously examined the coevolutionary interactions between plants (Brassicales) and butterflies (Pieridae), and uncovered evidence for an escalating evolutionary arms-race. Although gradual changes in trait complexity appear to have been facilitated by allelic turnover, key innovations are associated with gene and genome duplications. We are currently buliding upon these findings provide by investigating the specific functional performance and evolutionary dynamics of detoxification genes in the butterfly Pieris napi and relatives.
Principal Investigator: Christopher Wheat
Contributing Researcher: Kalle Tunstrom



