Julian Marius LudäscherPhD student
Research
Structure-based drug development targeting cancer
Cancer is an enormous challenge to treat and the progression of the disease results in significant limitations to quality of life, and even death [1]. Although cancer is associated with increasing age, it affects all age groups. For example, an analysis of global cancer statistics from 1990 to 2019 shows a slight overall increase in cancer among younger age groups [2]. While there has been a decrease in cancer-associated mortality due to the development of more advanced diagnostics and treatment options, the disease is still a major global public health problem and the development of therapeutic approaches is still a life-saving necessity.
A promising therapeutic approach for treating cancer is to target proteins involved in nucleotide metabolism [3]. Cancer cells grow at a faster rate than healthy cells, which they achieve through upregulating their primary metabolism. The excessive use of the respiratory chain then leads to increased formation of reactive oxygen species, which in turn damages DNA bases, ultimately leading to mutations. In order to avoid and remove these base lesions, metabolic nucleotide pathways are upregulated and therefore become essential for cancer cells. Inhibition of key proteins involved in nucleotide metabolism leads to the interruption of the respective pathway, and as a result cancer cells die due to increased mutations [4, 5].
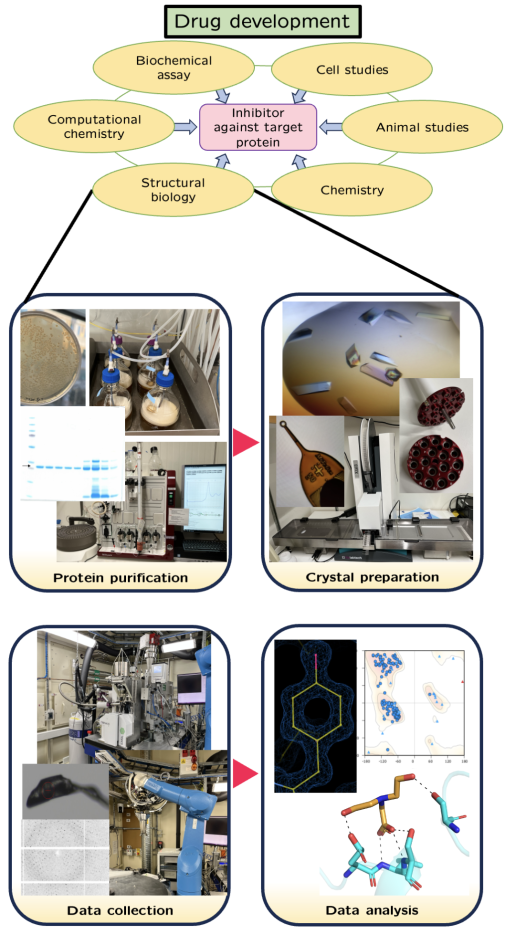
Effective inhibitor development from a starter molecule requires a combination of X-ray crystallography and enzyme activity studies. The focus of my PhD is on structure-based drug design to find and improve inhibitors targeting enzymes of nucleotide metabolism. To find potential inhibitors, I perform a comprehensive screening of fragment compound libraries. This is done by crystallographic fragment screening at synchrotron facilities. By obtaining protein–inhibitor complex structures from a screening, the binding of potential inhibitors can be analysed. The inhibitors are then optimised within an interdisciplinary project in order to improve their inhibitory effect. As soon as a potent inhibitor is created, it can be tested in clinical trials for final drug authorisation.
References:
1. Zugazagoitia J., Guedes C., Ponce S., Ferrer I., Molina-Pinelo S., Paz-Ares L. (2016). Current Challenges in Cancer Treatment. Clin Ther. 38, 1551-1566
2. You L., Lv Z., Li C., Ye W., Zhou Y., Jin J., Han Q. (2021). Worldwide cancer statistics of adolescents and young adults in 2019: a systematic analysis of the Global Burden of Disease Study 2019. ESMO Open. 6, 100255
3. Visnes T., Grube M., Hanna B. M. F., Benitez-Buelga C., Cázares-Körner A., Helleday T. (2018). Targeting BER enzymes in cancer therapy. DNA Repair. 71, 118-126
4. Carter M., Jemth A. S., Hagenkort A., Page B. D., Gustafsson R., Griese J. J., Gad H., Valerie N. C., Desroses M., Boström J., Warpman Berglund U., Helleday T., Stenmark P. (2015). Crystal structure, biochemical and cellular activities demonstrate separate functions of MTH1 and MTH2. Nat Commun 6, 7871
5. Gustafsson R., Jemth A. S., Gustafsson N. M., Färnegårdh K., Loseva O., Wiita E., Bonagas N., Dahllund L., Llona-Minguez S., Häggblad M., Henriksson M., Andersson Y., Homan E., Helleday T., Stenmark P. (2017). Crystal structure of the emerging cancer target MTHFD2 in complex with a substrate-based inhibitor. Cancer Res. 77, 937–948
$presentationText

