Integrative structural biochemistry
Evolution works by selection of point mutations, but precisely how do these point mutations affect the molecular behavior of the produced proteins? You will learn this during the course!
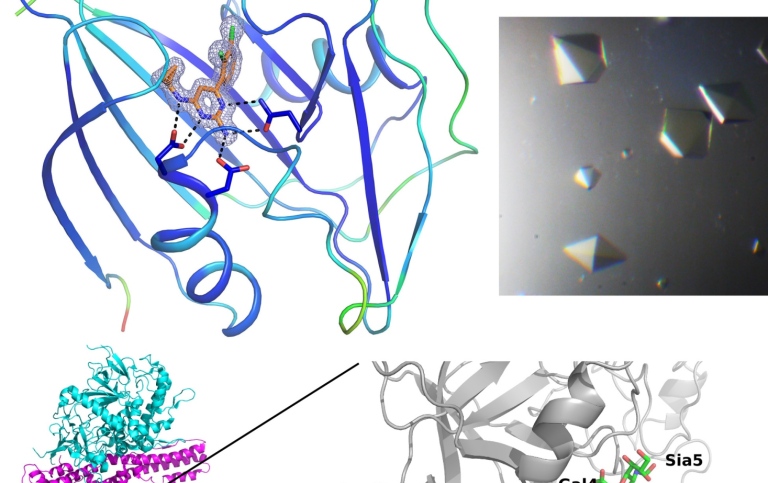
Evolution starts by the random and natural generation of point mutations in protein encoding genes. They can be silent and they can be loud and they are a natural consequence of replication in any organism where only favorable mutations are retained in the organism’s progeny. Depending on where these mutations are located in a protein’s three-dimensional structure they may lead to consequences for i.e. its folding, its activity, its intracellular interactions, its longevity in the cell and even a specific phenotype.
The function of a protein is tightly connected to its three-dimensional structure which can be determined using X-ray crystallography. In this course you will learn by doing.
You will learn how to determine a protein’s three-dimensional structure using X-ray crystallography structures from purified protein and subsequently explore how the protein’s property and function respond to point mutations.
The central focus is to understand how the detailed amino-acid interactions determine protein structural stability and binding. In this course you will be introduced to how theory, wet-labs and computing go hand-in-hand to solve real problems in protein chemistry.
In addition, application of basic chemical models and data quantification constitute a red thread throughout the teaching, and several common spectroscopic methods and experimental approaches are employed in depth.
Experimental results, progress and student conclusions will be presented/examined both in form of individual seminars and poster presentations.
-
Course structure
The course deals with the structure and properties of proteins, with a focus on sequence content and the effect of mutations. During the course you will learn about the following:
- The ability for reductionist thinking and generalization
- Quantitative description
- Formulation and testing of hypotheses according to falsification
- Proficiency in using laboratory techniques for problem solving
- Proficiency in computer-based methods and bioinformatics.
Expected learning outcomes
After completing the course the student is expected to be able to:- Explain the molecular principles behind the structure of proteins
- Describe entropy - enthalpy compensation in macromolecular systems and how it controls stability and structural properties
- explain mutation effects on protein stability and function, as well as the principles of structural evolution
- Formulate and test hypotheses about the structure, stability and function of proteins, as well as demonstrate proficiency in quantitative description and interpretation of experimental results
- Independently solve problems through the design of equilibrium experiments.
Modules
- Theory, 5 ECTS
- Laboratory exercises, 2.5 ECTS
Teaching format
- Lectures
- Group projects
- Seminars
- Lab work
Assessment
Theory - thorugh a written exam
Laboratory exercises - written and oral reports, mini conference presentation
Examiner
Ville Kaila
ville.kaila@dbb.su.se -
Contact
Course coordinator and examinerVille KailaProfessor
 Chemistry Section & Student Affairs Office
Chemistry Section & Student Affairs Office- Visiting address
Arrhenius laboratory, room M345
Svante Arrhenius väg 16 A-D
- Here you will find:
Student administrator
International coordinator
Study advisor
Director of studies
- Office hours
Monday, Tuesday and Wednesday 09.00-11.30 and 12.30-15.00
- Phone hours
Wednesday 10.00-11.30 and 12.30-15.00








