Induction and repair of DNA double strand breaks (DSBs)
One project in this laboratory focuses on how spontaneous DSBs arise and how they are repaired. The long-term goals are to create a better understanding of these processes to improve cancer treatment and to gain insights into the evolutionary process. For these studies, we use unicellular yeasts as model systems, namely the baker’s yeast Saccharomyces cerevisiae, and the milk yeast Kluyveromyces lactis. A vast array of molecular genetic tools is available for these organisms, making it possible to manipulate them for your own needs. It is known that basic cellular processes, such as DSB-repair and regulation of chromatin structure are conserved among eukaryotes. Together, this makes yeasts ideally suited for these studies.
Our recent work has focused on spontaneous DSBs arising during vegetative growth. We have found that in K. lactis, such breaks preferentially occur in transcriptional regulatory regions, suggesting a connection between transcription/ chromatin opening and DSB induction. DSBs are either repaired by NHEJ or HR and the two yeasts we have chosen to work with complements each other nicely in this aspect as K. lactis efficiently use NHEJ, whereas S. cerevisiae prefers HR.
Another recent discovery is mutant K. lactis strains that perform a process called mating type switch with dramatically increased frequency. Wild type laboratory strain perform mating type switch very seldom whereas these mutant strains appear to switch frequently. Mating type switch in K. lactis has not been explored before, but it is clear that this process differs significantly from the corresponding process in S. cerevisiae. Hence, learning about the mechanism and regulation of this process in K. lactis promises to be very interesting.
We also perform telomere studies in K. lactis. Telomeres are nucleoprotein complexes at chromosome termini that help to preserve overall genomic stability, in part by differentiating chromosome ends from DNA DSBs. As a result, DNA-repair activities are effectively inhibited at telomeres and within immediately adjacent chromosomal regions. Surprisingly, several proteins essential for NHEJ and HR in yeast are also involved in telomere maintenance and protection. This presents a paradox because in essence, the same proteins contribute to functionally opposing processes depending upon the genomic context. We therefore find it interesting to study the relationship between DSB-repair pathways and telomeres, in order to gain insight into this seeming dilemma.
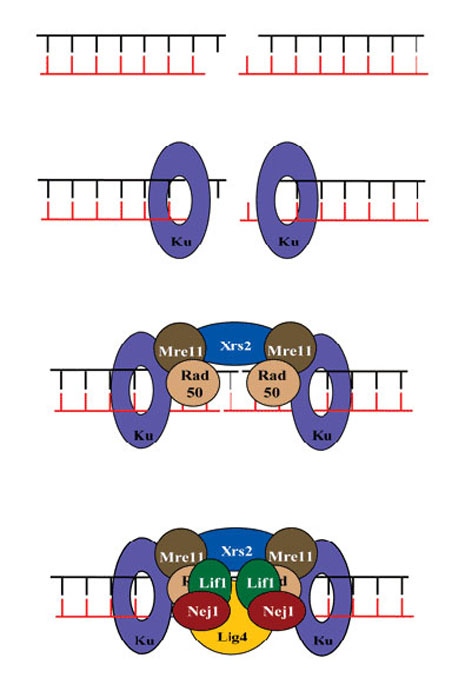
Molecular requirements for NHEJ
Ku70/80 binds the DSB with high affinity protecting the break from degradation and aligning the ends for the subsequent ligation reaction performed by the Lig4/Lif1/Nej1 complex. The MRX-complex (Mre11, Rad50 and Xrs2) are thought to form a molecular bridge between the two ends, ensuring efficient repair.
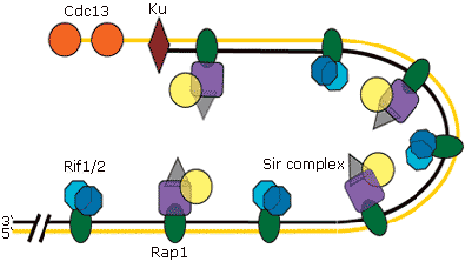
The telomere cap in yeast
Telomeric repeats contain specific binding sites for Rap1, which recruits the Sir-complex and Rif1/Rif2 to the chromsome ends. In addition, the NHEJ proteins Ku70/80 binds telomeres. The single stranded overhangs are bound by Cdc13. Loss of the telomere cap leads to that telomeres are considered to be DSBs. The DSB-repair machinery thus attempts to repair the telomeres by NHEJ or homologous recombination. Such repair events may result in chromsome fusions, commonly observed in cancers.




