Research group Group Ljungdahl
Amino acids are essential nutrients that serve as building blocks of proteins and some are efficiently metabolized for energy. Eukaryotic cells respond to extracellular amino acids by enhancing their uptake. We study the molecular mechanisms underlying this response and the role of amino acid metabolism in promoting virulent growth of human fungal pathogens.
Group description
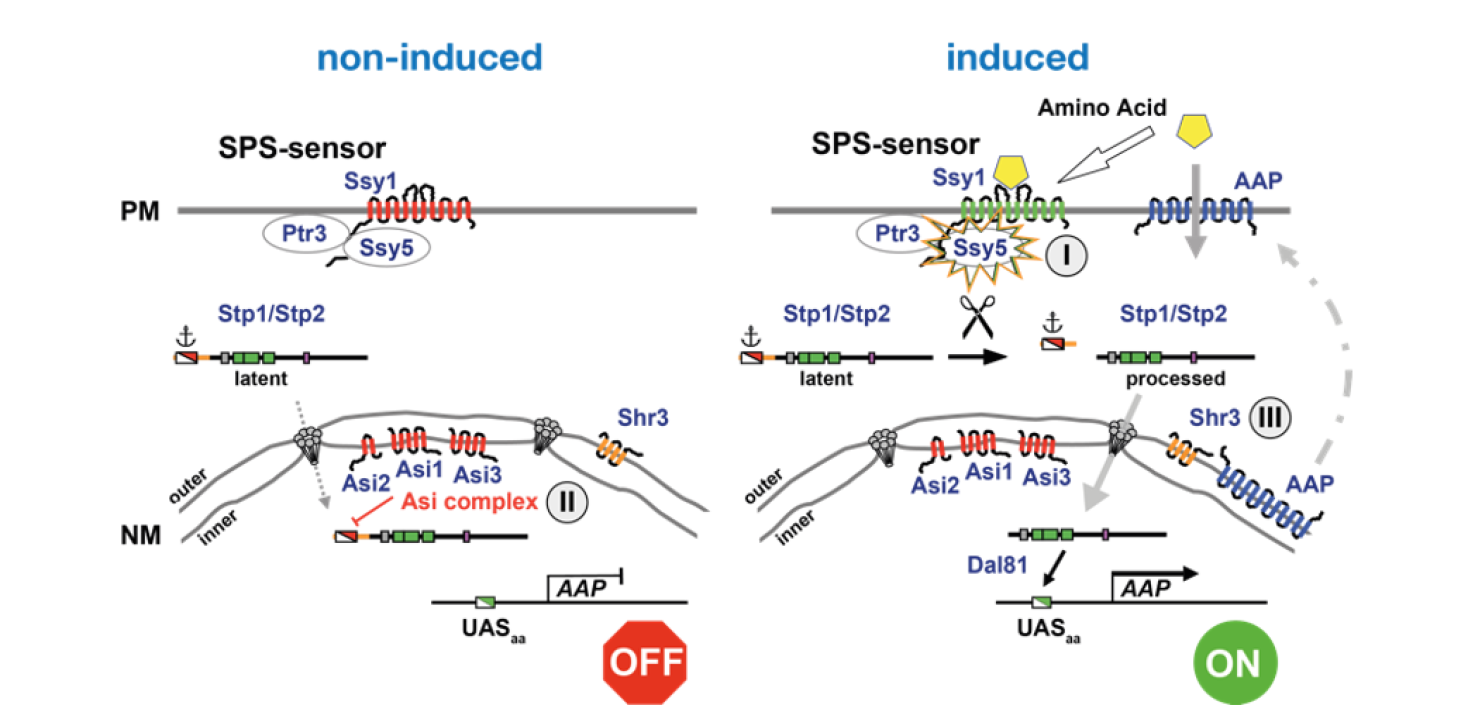
Nutrient uptake is essential for cellular life and the capacity of cells to perceive extracellular nutrients is critical for coordinating their uptake and metabolism. The analysis of nutrient-regulated gene expression remains a substantial challenge in contemporary biology, and deciphering the entire repertoire of signaling mechanisms is a requisite to understand how cells properly control growth and development. Yeast and mammalian cells respond to the presence of low levels of extracellular amino acids by enhancing their uptake. We are studying the molecular mechanisms underlying this in the model yeast Saccharomyces cerevisiae. The use of yeast has allowed us to rationally investigate the mechanisms of signal transduction in great detail. We have defined the key signaling events that define the SPS-sensing pathway, and have achieved a general understanding of how amino acid-induced signals are transduced from the plasma membrane to the promoters of responsive genes (see Figure 1). Our results have revealed several novel eukaryotic-specific regulatory mechansims that are spatially separated in cells. Specifically, we have defined the trimeric SPS sensor (Ssy1-Ptr3-Ssy5) that initiates amino acid-induced signaling events at the plasma membrane, the Asi-E3 ubiquitin ligase complex (Asi1-Asi2-Asi3) localized to the inner nuclear membrane that is essential to maintain the “OFF-state” of SPS-sensor regulated genes, and highly specific endoplasmic reticulum (ER) membrane-localized chaperones that function to prevent inappropriate molecular interactions between hydrophobic segments of polytopic membrane proteins as their membrane-spanning segments insert and partition into the ER membrane during synthesis. Although these findings are in seemingly disparate areas of research, i.e., signal transduction, protein degradation and membrane protein biogenesis, our results are in fact highly interrelated. The major amino acid sensor and signaling component in the plasma membrane Ssy1, and the proteins catalyzing amino acid uptake are related and comprised of twelve membrane-spanning segments and mutually require the membrane-localized chaperone Shr3 for functional expression. Our results provide a clear example that a broad understanding of cell biology is required to fully understand cellular signaling systems.
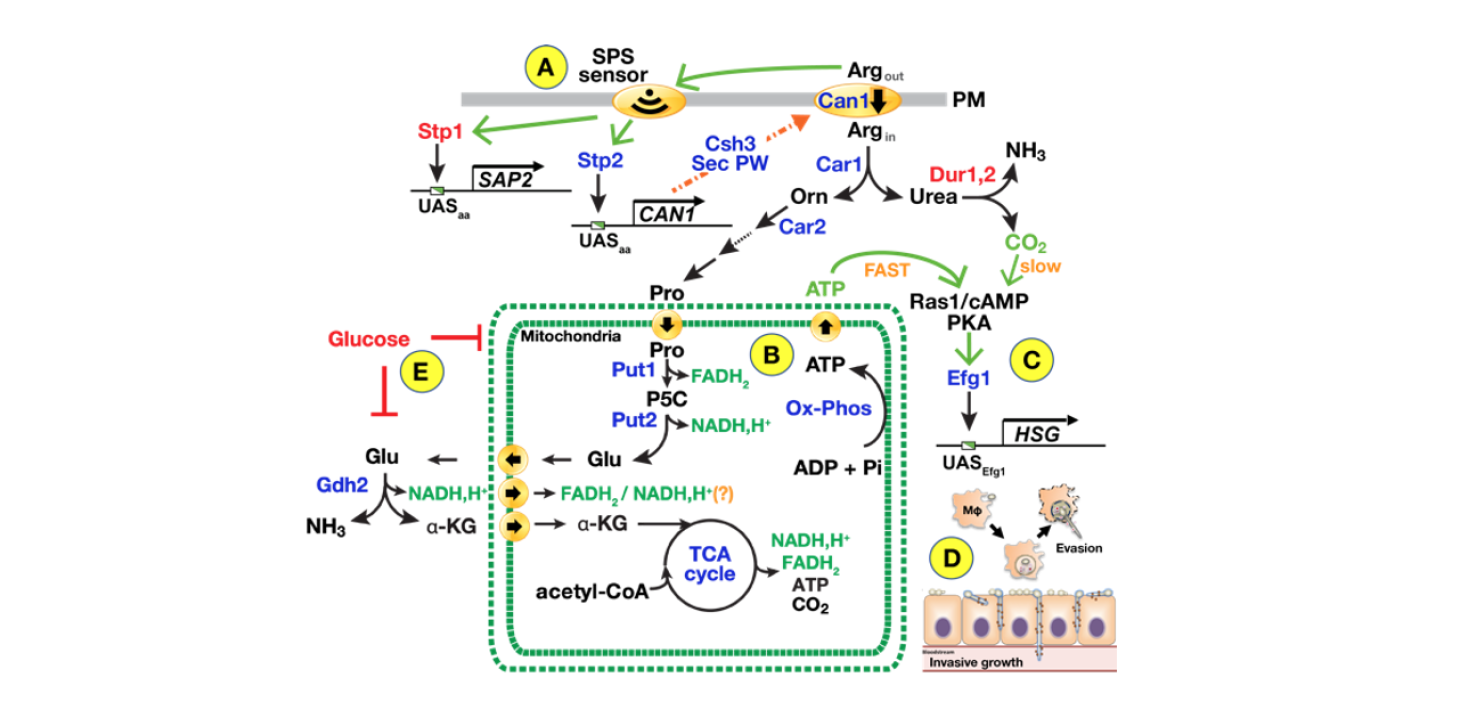
Humans are host to a wide spectrum of commensal microorganisms collectively known as the human microflora. The microflora is largely composed of prokaryotic bacteria, however, eukaryotic fungi are also major components, with Candida spp. dominating. Many species of Candida, e.g., Candida albicans, Candida glabrata and Candida auris, are opportunistic pathogens that can cause life threating infections in immune compromised individuals. As the incidence of candidiasis is quite low in healthy populations, environmental factors, such as interactions with the primary immune cells play critical roles. Our studies in yeast have established paradigms to understand nutrient-regulated processes in Candida spp. However, fundamental and important differences exist. In constrast to yeast, which evolved in high-glucose environments and can readily be found in nature, Candida spp. have evolved in close association with human hosts and are not found living freely in nature. Candida spp. are well-adapted for growth in the low glucose environment of human hosts and efficently metabolize amino acids as energy sources. There are high levels of amino acids in circulateing blood, and host proteins are rich sources of amino acids that can be liberated by proteases that cleave them into smaller peptides and free amino acids. The advent of CRISPR/Cas9-based methods have facilitated the genetic analysis of human fungal pathogens and we are applying state-of-the-art molecular biological techniques to directly examine growth requirements in vivo and in situ in infected model mammalian hosts. We have recently identified mitochondrial proline catabolism as critical for inducing and energizing filamentous growth, a virulence feature that underlies evasion from macrophages and the ability to invade across endo- and epithelial barriers (see Figure 2). Building on this knowledge, we are pursuing three aims: 1) Fully characterize the metabolic control of proline-dependent fungal virulence, specifically the role of mitochondrial-localized processes that are critical to fungal cell survival in hosts; 2) Visualize the spatio-temporal aspects of C. albicans infections in the kidney of a living mammalian host and define host-pathogen interactions using advanced intravital 2-photon and STED microscopy and spatio-transcriptomic analysis; and 3) Define the virulence properties of multidrug resistant Candida auris. We anticipate that the results will provide a solid foundation for developing novel therapeutic strategies in the expanding population of immune compromised individuals.
Keywords: Saccharomyces cerevisiae, Candida albicans, Candida glabrata, Candida auris, nutrient sensing, signal transduction, polytopic membrane protein biogenesis, membrane-localized chaperone, proline metabolism, mitochondria
Group members
Group managers
Per Ljungdahl
Professor

Members
Marisol Torres Gonçalves
Geust PhD student

Sofia Dimou
Postdoc

Viktoriia Tsuber
Forskare

Marzia Rizzo
Forskare

Maryam Nejati
Forskare

Ioanna Myronidi
Doktorand