Epigenetics can be defined as non-genetic changes that are transmitted through cell-divisions. The purpose of this project is to determine the function of chromatin modifying proteins in epigenetic inheritance during animal development. Chromatin modifications, such as histone acetylation and methylation may constitute an epigenetic code that influences the transcriptional state of the genome.
Epigenetic inheritance in Drosophila
There are two well-studied epigenetic phenomena in Drosophila melanogaster. The first is position-effect variegation (PEV), which results in the clonal silencing of genes juxtaposed to heterochromatin. Genetic screens identified suppressors and enhancers of PEV, which has provided insights into the molecular processes causing heterochromatic gene silencing (Ebert et al. 2006). In particular, with the demonstration that Suppressor of variegation 3-9, Su(var)3-9, is a histone methyltransferase responsible for histone 3 lysine 9 (H3K9) methylation, and that Su(var)2-5 encodes heterochromatin protein 1 (HP1), a key step in heterchromatin formation was identified.
The second epigenetic process involves Polycomb group (PcG) and trithorax group (trxG) proteins. PcG proteins are evolutionarily conserved chromatin regulators that epigenetically maintain appropriate expression patterns of developmental control genes, such as the Hox genes, when the original transcriptional activators and repressors are no longer present (Ringrose and Paro 2004). PcG proteins are generally repressors that maintain the off state of genes, and exist in at least two distinct protein complexes. The Esc-E(z) complex is a H3K27 specific histone methyltransferase. Polycomb repressive complex 1 (PRC1) can block chromatin remodeling by the SWI/SNF complex in vitro. The PRC1 subunit dRing1/Sce, as well as its mammalian orthologs, are E3 ubiquitin ligases that monoubiquitylate histone H2A, which is necessary for Hox gene repression.
TrxG proteins are required to maintain appropriate levels of Hox gene expression during embryogenesis, and counteract the effects of PcG genes (Ringrose and Paro 2004). Several trxG proteins are constituents of SWI/SNF-type chromatin remodeling complexes, and some are H3K4 methyltransferases,
Drosophila Reptin and the TIP60 complex in epigenetic inheritance of silent chromatin
Reptin and the related Pontin protein are DNA helicases present in different chromatin remodeling complexes. In Drosophila, Reptin is part of the TIP60 HAT complex, but also co-fractionates with the PcG complex PRC1 (Saurin et al. 2001). We have examined the role of Drosophila Reptin in chromatin regulation. We found that reptin mutant flies genetically interact with PcG genes to prevent mis-expression of the homeotic gene Sex combs reduced (Scr) in leg imaginal discs. In wing imaginal discs containing homozygous mutant reptin clones, generated by FLP-FRT-mediated mitotic recombination, the homeotic gene Ultrabithorax (Ubx) is not ectopically expressed, whereas it is in clones generated from most PcG mutants. From these experiments, we conclude that Reptin does not behave as a bona fide PcG gene that is able to de-repress homeotic gene expression, but rather cooperates with PcG proteins to silence genes.
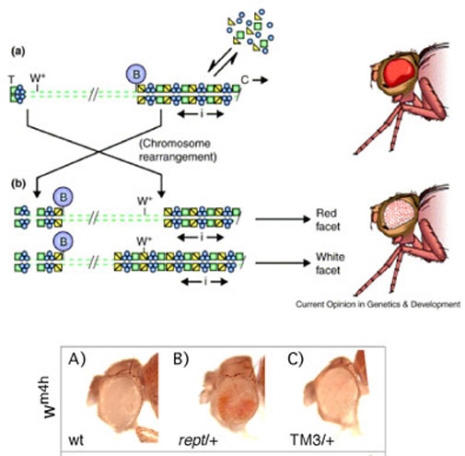
We then tested if Reptin has a role in another epigenetic process, position-effect variegation (PEV), and found that reptin mutants suppress PEV at many different loci(Fig 1). This means that Reptin normally contributes to the spreading of heterochromatin. Since genetic interactions with PcG genes and suppression of PEV are features shared with two other components of the TIP60 complex, namely Enhancer of Polycomb and Domino, we characterized mutations in yet another TIP60 component, dMRG15. We found that dMRG15 mutants also suppress PEV and interact with PcG genes. Based on these results, we suggest that Drosophila Reptin is acting through a fly TIP60 complex to control epigenetic processes leading to a repressive chromatin state (Qi et al. 2006).
In future experiments, we will determine whether the enzymatic activities associated with the TIP60 complex, histone acetylation and histone variant exchange, contribute to epigenetic gene regulation.
Histone lysine demethylases
Histone lysine methylation has been regarded as a stable chromatin modification that together with DNA methylation mediates epigenetic inheritance. The recent discovery of histone lysine demethylases indicates that these epigenetic marks can be reversed under some circumstances (Shi and Whetstine 2007). A family of proteins containing Jumonji C (JmjC) domains was recently proposed to encode histone demethylases (Shi and Whetstine 2007). By database searches we identified 13 JmjC-containing genes in Drosophila, and found lethal transposon insertions in four of them. We will investigate the contribution of these gene-products to epigenetic inheritance by assaying their effect on PEV, and by looking for genetic interactions with PcG and trxG genes. Preliminary results indicate that some of them, including mutations in little imaginal discs (lid), enhance PcG phenotypes. We have subcloned 9 out of the 13 cDNAs into cell-culture expression vectors and will assay their effect on global histone methylation when over-expressed. This far, we have found that Lid over-expression reduces H3K4 tri-methylation. Since methylation of H3K4 by the trxG proteins Trithorax and Ash1 prevents silencing by PcG proteins, our intriguing results (interaction with PcG genes and H3K4 demethylation) indicate that PcG proteins may counteract TrxG function in part by Lid-mediated H3K4 demethylation.
References
Ebert, A., S. Lein, G. Schotta, and G. Reuter, Histone modification and the control of heterochromatic gene silencing in Drosophila. Chromosome Res. 2006. 14: 377-92.
Qi, D., H. Jin, T. Lilja, and M. Mannervik, Drosophila Reptin and other TIP60 complex components promote generation of silent chromatin. Genetics. 2006. 174: 241-51.
Ringrose, L. and R. Paro, Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet. 2004. 38: 413-43.
Saurin, A.J., et al., A Drosophila Polycomb group complex includes Zeste and dTAFII proteins. Nature. 2001. 412: 655-60.
Shi, Y. and J.R. Whetstine, Dynamic regulation of histone lysine methylation by demethylases. Mol Cell. 2007. 25: 1-14.




