Research project Establishment of epigenetic landscapes
Epigenetic patterns of histone modifications contribute to the maintenance of tissue-specific gene expression, but little is known about how such patterns are initially formed during early embryo development. We study this process and how cell type-specific patterns emerge in Drosophila.
We investigate how the three germlayers mesoderm, neuroectoderm, and dorsal etoderm come to differ in their epigenetic patterns. Single cell experiments and mutants in the Toll signaling pathway are used to study different cell types, and genomic methods such as single cell RNA-seq, single cell CUT&Tag, ChIP-seq, ATAC-seq, Hi-C, and PRO-seq are applied to follow how histone modifications, chromatin accessibility, chromatin conformation and transcription are established in the different cell types. Our studies address how transcriptional enhancers communicate with promoters, and how the chromatin state influences this communication and transcriptional output.
Project description
The early Drosophila embryo is patterned along the anterior-posterior and dorsal-ventral axes by transcription of developmental control genes in different parts of the embryo. Dorsal-ventral patterning is controlled by an intra-nuclear concentration gradient of Dorsal, a Rel-family transcription factor related to NF-kappaB. Over 50 Dorsal target genes are known, and this gene regulatory network constitutes one of the best understood in the development of any animal.
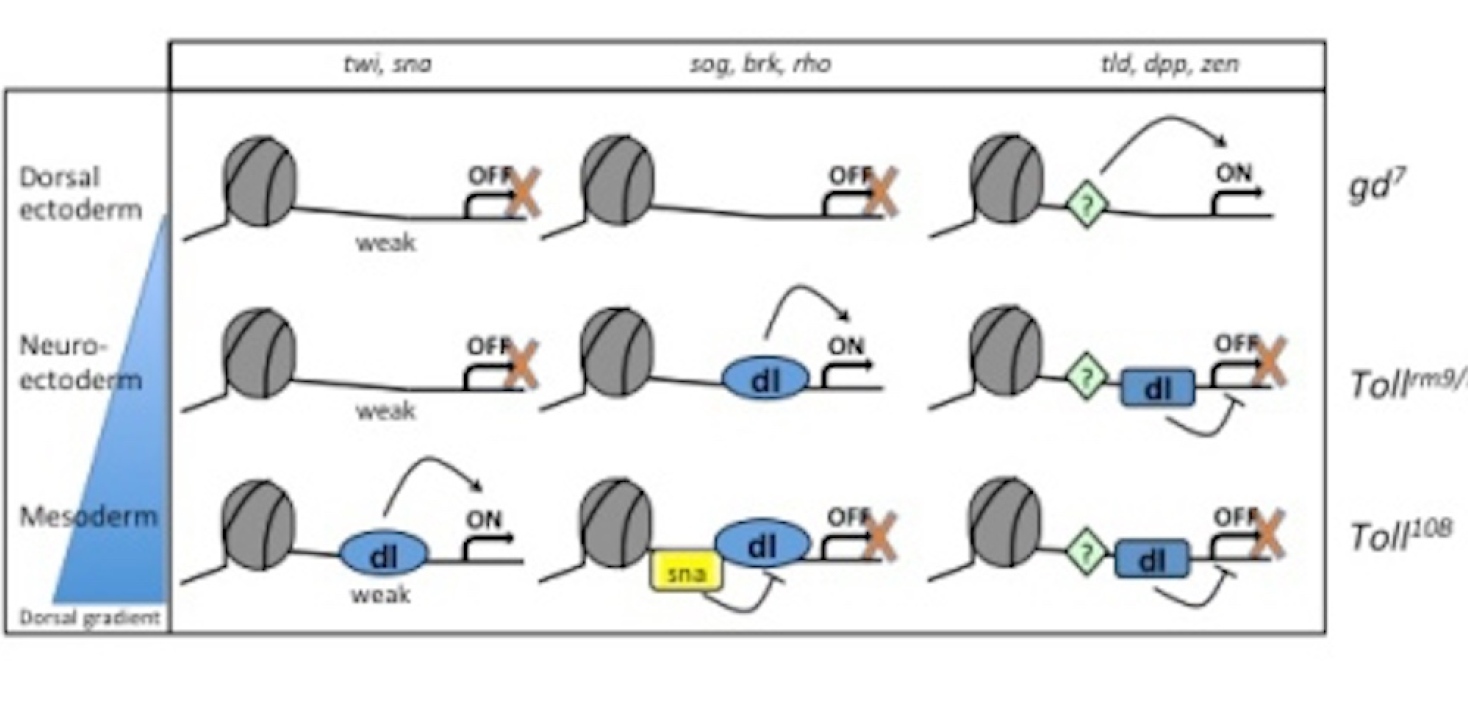
Investigation of dorsal-ventral patterning in Drosophila provides a unique opportunity to study cell specification, since mutants in the Toll signaling pathway can be used to generate embryos with a homogenous population of cell types (see Fig. 1). In embryos derived from gd7mutant mothers, Dorsal fails to enter the nucleus and the entire embryo is transformed into dorsal ectoderm. In embryos derived from Tollrm9/rm10 mothers, all cells become neurogenic ectoderm since Dorsal uniformly enters nuclei at an intermediate concentration. Toll10Bmothers produce embryos where Dorsal enters all nuclei at high concentration thereby transforming them into mesoderm. Chromatin extracts from these mutant embryos circumvent the problem of a mixed population of cell types, but enable investigation of histone modifications and regulator binding to endogenous loci under conditions that the different cell types normally experience during development. These tools allow us to study mechanisms of tissue-specific gene expression in vivo, in a manner that is exclusive to the early Drosophila embryo.
We have found that the epigenetic landscape at two neuroectoderm genes, brk and sog, differ between the three gemlayers (Boija and Mannervik 2016). These loci are decorated with Polycomb-mediated H3K27me3 in the dorsal ectoderm where they are off, have H3K27ac in the neuroectoderm where they are on, and lack both modifications in the mesoderm where they are repressed by Snail (Boija and Mannervik 2016). Using PRO-seq, ChIP-seq, CUT&Tag, and ATAC-seq we have characterized how the chromatin gradually gains accessibility and histone modifications at all dorso-ventral genes as the cells become specified. This results in release of a promoter-proximal paused Pol II that is established already in naïve cells in the cell type where the gene is expressed (Hunt et al. 2024) Interestingly, despite differences in chromatin state and transcription between tissues, the three-dimensional genome architecture does not differ between the three germlayers (Ing-Simmons et al. 2021). This shows that differential gene expression is independent of 3D chromatin conformation in the early embryo. Current work investigates how transcriptional enhancers communicate with promoters without changes in chromatin conformation, and how the chromatin state influences this communication and transcriptional bursting.
Using a single cell CUT&Tag method, nano-CUT&Tag, we have profiled the active H3K27ac mark and the repressive H3K27me3 modification simultaneously in cells at later developmental stages. Using this data we are modelling the epigenetic landscape and how it influences embryo development.

